Cross- functional teams from science, logistics and sample handling, operations, and project management engage early on. This ensures we can secure your critical clinical endpoints.

We have central laboratories in the USA, Europe, Asia Pacific, China and Australia. This means you can count on Cerba Research to handle study set up, site preparation and sample collection, through to delivery. Results are reported via our single global database, providing the end point reporting you need to keep your clinical trial on track.
More than providers of specialty and central laboratory testing, our team has years of experience in shaping testing protocols and patient cohorts to help secure successful outcomes. Our experience becomes your expertise.
Collaboration is key and we understand that people matter. Whether you have detailed scientific questions, or want to know more about how we can deliver, we have experts in science, clinical trial operations , and project management that speak your language. From safety testing to PMBC & BMMC, Cerba Research’s knowledge spans all aspects of central lab research.
We can advise early on how to approach the selection and set up of your global central laboratory. Unfortunately, often sponsors leave the central and specialty laboratory considerations until later in their outsourcing processes. Our long standing clinical experience demonstrates that early engagement leads to better protocol insights and more efficient data collection.

Our team of dedicated scientists will work with you to understand your needs from preclinical through to clinical trials and tailor assay development to meet your research, clinical or commercial objectives.

With our unparalleled scientific expertise, the fast-expanding portfolio of therapeutic areas includes a focus on oncology and immuno-oncology, metabolic diseases, virology, infectious diseases, inflammatory and autoimmune diseases, and cell and gene therapy.
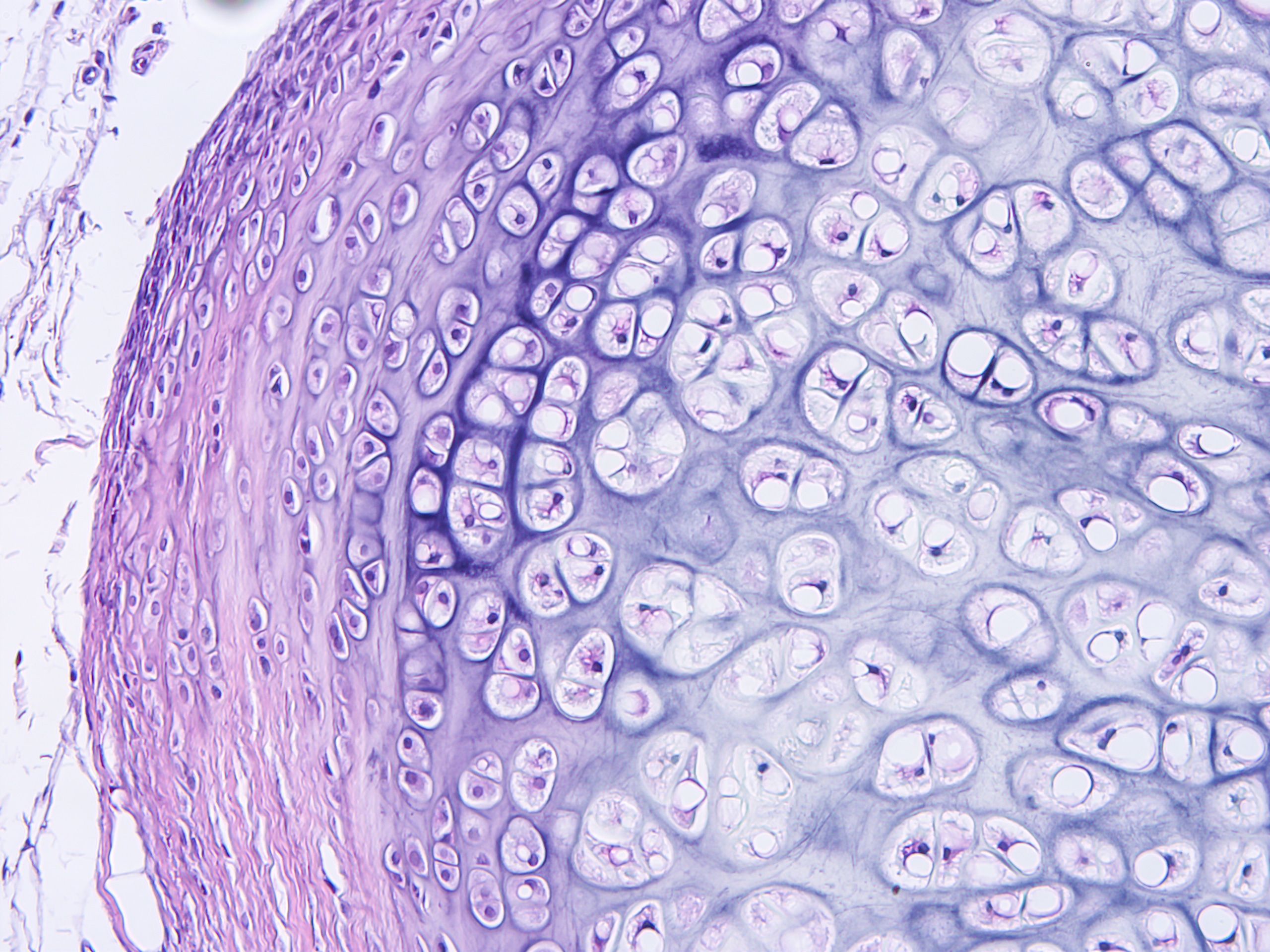
Cerba Research has an extensive history providing specialty lab and biomarker services. It’s in our DNA. We can help you optimize your protocol development through early scientific insights, and our next generation assays can provide a targeted or broad approach to immune profiling through our multifactorial biomarkers. Benefit from our customised service to obtain innovative, flexible solutions grounded in expert science.
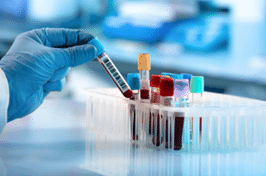
Our global network of pre processing laboratories ensures we are ideally suited to isolate and either store and analyze PBMC and BMMC samples on a global scale. We guarantee customers a high level of quality from sample collection to processing, shipping, handling, storage and analysis worldwide.
We meticulously monitor laboratory results with daily instrument calibrations and an extensive internal quality control program. National certifications such as CLIA, CAP, and ISO are indicative of stringent quality management in all aspects of our organization.

Reach out to our experts and see how we can help advance your clinical trial
Contact Us